Portfolio

LinkSēq - qPCR
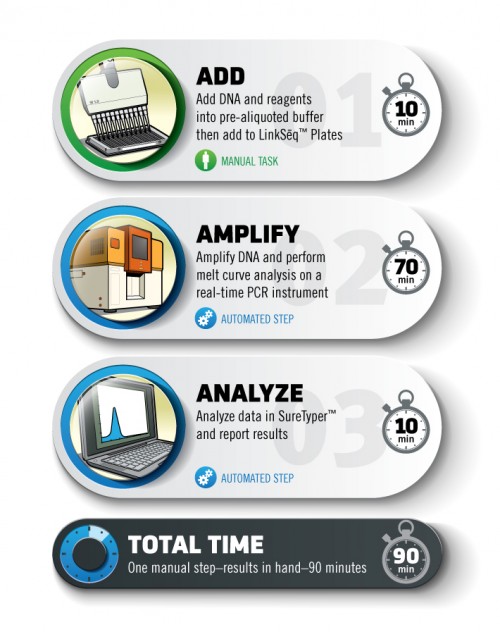
With less than 10 minutes of set-up and no further operator intervention, LinkSēq™ uses state of the art real-time PCR detection to provide complete molecular genotyping results in approximately 90 minutes.
- More productivity with less effort
- Fewer repeats with less steps and reproducible results
- Accurate results with automated analysis
Innovative Technology
The LinkSēq™ methodology is based on allele specific PCR similar to current SSP methods; however, unlike SSP, LinkSēq utilizes SYBR® Green to identify the presence or absence of alleles.
- Unique Real-Time PCR based genotyping test
- No gels (SSP)
- No hybridization or washing steps (SSO)
- Designed to provide digital Yes or No results
Melt Curve Analysis
LinkSēq™ uses SYBR® Green, a non-toxic alternative to ethidium bromide, to determine the presence or absence of specific alleles. When bound to double stranded DNA, SYBR Green fluoresces. A Real-Time PCR instrument is used to take fluorescent readings of each well at different temperatures. As the temperature increases, the DNA will “melt” and the fluorescence will decrease. Graphing the melt-curve makes visualizing the results easy, and allows the software to easily identify if an allele is present or absent.

- No Ethidium Bromide
- Each Assay is either positive or negative (allele is present or absent)
- SureTyper software utilizes the expected melt temperature for each reaction
- Accurate typings a result of robust chemistry
Automated Results
SureTyper™ is the software platform used in combination with LinkSēq™ Real-Time PCR kits for automating data interpretation and simplifying genotype reporting.
- Easy to Use Interface
- Automated Data Interpretation and Typing Analysis
- Examination of Raw Data
- Flexible Reporting Options
LinkSēq Benefits
LinkSēq is a rapid genotyping method with a simple workflow that avoids post-amplification contamination. Given the ease of use, rapid turn around times, and increased laboratory productivity, LinkSēq offers an attractive improvement over current SSP and SSOP typing methods.
- Increased productivity
- No contamination control needed
- No gels to pour or gel boxes to clean
- No washing, probing, or transfer of PCR products
- Reduced analysis effort from built in redundancy and design features
- Only seconds to call typing assignments


